USP Chapter Covers Residual DNA Testing Practices For CHO And E. coli
By Shubrata Khedkar and Bruno De Carvalho, United States Pharmacopeia

Biomanufacturers often use bacterial, yeast, insect, and mammalian cell lines to support the production of biological therapy products.1 Each cell type has its own benefits and drawbacks and biomanufacturers select the cell type based on the requirements of the therapeutic product.
Mammalian Chinese hamster ovary (CHO) cells are widely used for the commercial production of biological medicines since they can be cultured in suspension, allowing volumetric scalability and use of large stirred-tank bioreactors.2 Such manufacturing platforms come with the drawback of carrying impurities such as host cell proteins and host cell DNA. The resulting drug substances must be carefully monitored, and the downstream purification processes must be capable of removing these impurities. Residual host cell DNA is one of the most critical process-related impurities, posing significant safety risks due to its potential immunogenicity, infectivity, and oncogenicity.3 These impurities can lead to the transfer of viral DNA and possible genotoxicity from activated oncogenes in the cell substrates used for production. The oncogenicity of residual DNA is a major source of concern in platforms based on tumorigenic cell lines, such as HeLa cells. Additionally, there is a risk of aberrant gene expression by the insertion of residual DNA into the genome.4,5
Therefore, demonstrating that the level of residual DNA present in a biological product is below a certain criterion is important for ensuring product safety. The level of residual DNA is also an indication of the overall quality of the purification process.6 The commonly accepted limits are ≤10 ng of detectable residual DNA per dose7 and any persisting DNA must be ≤200 base pairs in the final product.8 But these limits can vary based on the risks associated with the product and its administration. The World Health Organization (WHO) recommends that manufacturers consider the characteristics of the cell substrate, the intended use, and the route of administration when setting residual DNA limits. Manufacturers should also assess the effect of the manufacturing process on the size, quantity, and biological activity of residual DNA fragments.7 However, to accomplish this, the manufacturer must have methods that can quantify residual DNA levels at different points throughout the manufacturing process.
Methods such as membrane hybridization and protein-based DNA binding assays traditionally have been used to quantify residual DNA, but these classic methods are time-consuming and have reproducibility issues.
A more reliable approach uses quantitative real-time PCR (qPCR) to monitor the amplification of a target residual DNA sequence. In the probe-based qPCR method, DNA probes labeled with a fluorescent reporter and quencher dye are designed to bind to a specific DNA sequence. During the PCR cycles, as a new strand is generated by DNA polymerase, this enzyme also cleaves the probe separating the reporter from the quencher and consequently generates fluorescence. The probes contain sequences complementary to a target residual DNA and generate a very specific signal for residual DNA tests.
USP performed a global survey to better understand the state of industry practices regarding residual DNA testing. The responses indicate that qPCR is the most used technique for this purpose due to its sensitivity, specificity, dynamic range, and high-throughput capability. Industry respondents also highlighted the availability of standardized best practices and the familiarity of regulators with qPCR. What was clearly lacking was a documentary standard that integrated all the information into a single source that manufacturers could leverage to develop and validate assays that would meet regulatory expectations. To address this need, USP collaborated with various stakeholders to develop the USP-NF General Chapter <509> Residual DNA Testing, which is intended to support the manufacturing and quality assessment of biological products.
The General Chapter focuses on E. coli and CHO cells because they are two of the most used cell host substrates for manufacturing recombinant proteins. CHO cells alone account for 60% to 70% of recombinant biologics on the market.1 A compendial solution with a validated assay for the measurement of residual DNA in CHO and E. coli systems is a critical tool to guarantee the quality of biological products.
The USP Solution For Residual DNA Testing: General Chapter <509>, Genomic DNA Reference Standards, and Analytical Reference Materials (ARMs)
USP began this work by gathering global industry and regulatory experts to create a USP Expert Panel. One of the first challenges they tackled was the reproducible extraction of DNA from high-concentration protein samples, typical of most biological products. Most samples require a pretreatment step to remove any interfering substances from the sample matrix before residual DNA can be measured. The simplest sample manipulation is dilution in water or Tris EDTA buffer to reduce matrix interference, provide an appropriate starting volume, and bring the analyte concentration within the quantitative range of the qPCR method.9 However, overcoming assay interference might require additional sample manipulation, including proteolytic digestion, chemical dissociation, or extraction.1
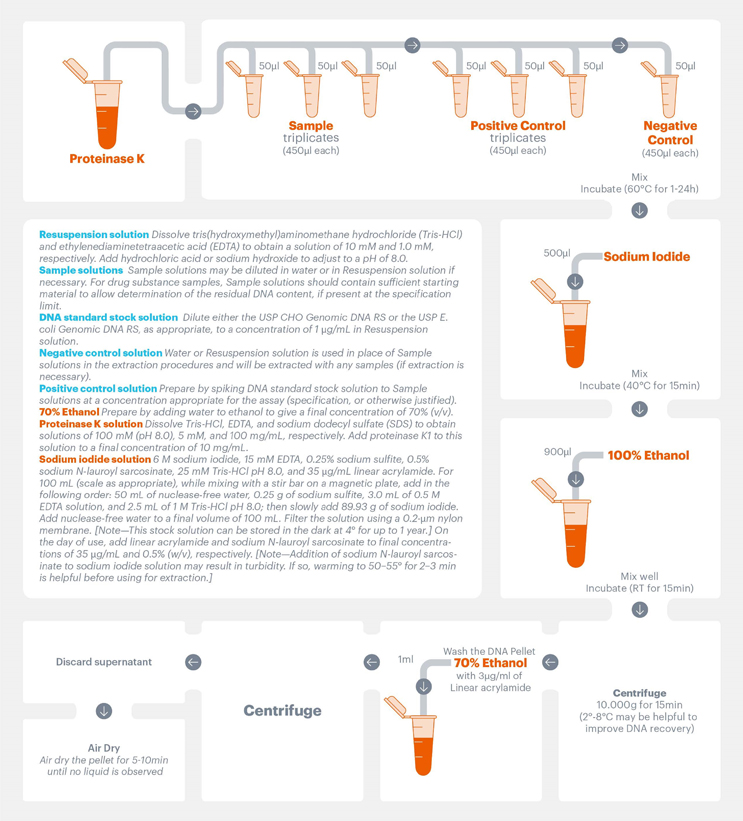
In our industry survey, almost all the respondents were using DNA extraction methods to prepare samples for assessment, and more than half were using chaotropic agents (e.g., sodium iodide). The USP General Chapter <509> presents sample preparation and extraction procedures that can be used prior to qPCR-based methods for the measurement of residual DNA. Extraction procedures covered in the General Chapter include those that do not require any specific commercial kits, such as Proteinase K digestion followed by sodium iodide extraction and ethanol precipitation. The extraction procedure was optimized and validated to accommodate starting DNA concentrations ranging from 0.01 to 50 pg/mL. An overview of the compendial extraction method is presented in Figure 1.
The qPCR method described within the General Chapter is extremely sensitive and specific for detecting residual genomic DNA. Primer and probe sequences are also provided, and the method allows for several different labeling strategies. In addition, system suitability criteria and sample acceptance criteria are described in detail; however, product-specific limits were not included since it is well known that, depending on product type, this value can vary and is negotiated between the manufacturer and regulators.
General Chapter <509> is supported by rigorously quantitated USP Reference Standards to measure E. coli genomic DNA (#1231557) and CHO genomic DNA (#113710) for products made in these cell substrates. The standards can be used to generate standard curves for routine assays, as a positive control to spike into samples for extraction, as a standard to monitor and trend method performance, or for analyst training. It is important to mention that the method specifications mentioned in this General Chapter apply to its designated USP reference standards only. The results obtained using non-compendial reference standards are considered inconclusive and would require a new method validation.
As part of a strategic collaboration with ATCC, USP also introduced genomic DNA Analytical Reference Materials (ARMs) for measurements of residual DNA. These materials support residual DNA detection for products developed in the host cells HEK-293 (#1592106), Sf9 (#1592170), MDCK (#1592111), MRC-5 (#1592112), Vero (#1292190), and BHK-21 (#1592100). Application notes detailing tests to evaluate the quality and applicability of each of these ARMs are published and accessible.
Figure 1. Schematic illustration of the extraction method described in the USP General Chapter <509>. Click to enlarge.
Compendial Methods Save Time, Reduce Costs, And Mitigate Risk
It is possible to find kits for residual DNA detection provided by commercial companies. A major issue associated with these non-compendial solutions is the need for validation. According to the FDA’s Analytical Procedures and Methods Validation for Drugs and Biologics Guidance for Industry,10 non-compendial methods require analytical validation, which tests attributes such as specificity, linearity, accuracy, and many others, which can add significant cost and time for a lab. It also requires highly trained personnel and there is a risk that the resulting method may not pass regulatory review. Compendial methods facilitate this process. It is still necessary to demonstrate the suitability of the compendial procedure for the drug product as described in the FDA’s guidance for industry and USP General Chapter <1226> Verification of Compendial Procedures. However, the method is validated, and no robustness studies must be included if methods are followed without deviation.8
Another advantage of using USP compendial standards is that they mitigate risks associated with the use of commercial kits, such as discontinuation of reagents or instruments. This risk is mitigated by USP’s “brand-agnostic” approach. For example, USP General Chapter <509> provides the sequence of the primers and probes needed for the compendial method. This information is usually not disclosed by commercial companies, setting a dynamic where users are prevented from ordering these oligonucleotides from other suppliers. Another example of the flexibility that compendial methods provide is the detailed description of each required reagent. As shown in Figure 1, users can prepare their own reagents without having to rely on commercial kits of unknown components that are often modified or discontinued with time. It is also known that standards present in these kits are not standards from a recognized authority such as WHO or a pharmacopeia, so additional resources may be required by the user to verify the value of these materials.11, 12 Therefore, a compendial solution for applications such as residual DNA detection benefits manufacturers of biological products.
Conclusions
Residual DNA is a critical quality attribute that must be controlled and carefully monitored in all biological therapies, including mAbs and protein therapies. There are strict limits imposed by regulatory authorities on the amount of residual DNA allowed in biological medicines. Companies still struggle with how to appropriately develop and validate assays that meet regulatory expectations. As a standards-setting organization, USP can address this need and has developed General Chapter <509> Residual DNA Testing to provide companies with best practices and validated procedures that enable them to develop their own assays for products produced in E. coli and CHO cells. The methods within the General Chapter were chosen after consulting industry participants and are the most tried and true procedures being implemented today. USP also provides ARMs to support residual DNA detection for products produced in HEK-293, Sf9, MDCK, MRC-5, Vero, and BHK-21 cells. In the future USP intends to release additional materials and standards supporting residual DNA testing. Industry and regulatory experts are encouraged to work with USP and other pharmacopeias to develop and advance methods for the detection of residual DNA in products using other host cells for production.
References:
- Zhu, M.M., et al., Industrial production of therapeutic proteins: cell lines, cell culture, and purification. Handbook of industrial chemistry and biotechnology, 2017: p. 1639-1669.
- Jayapal, K.P., et al., Recombinant protein therapeutics from CHO cells-20 years and counting. Chemical engineering progress, 2007. 103(10): p. 40.
- Wang, X., et al., Residual DNA analysis in biologics development: review of measurement and quantitation technologies and future directions. Biotechnology and Bioengineering, 2012. 109(2): p. 307-317.
- Sheng-Fowler, L., A.M. Lewis Jr, and K. Peden, Issues associated with residual cell-substrate DNA in viral vaccines. Biologicals, 2009. 37(3): p. 190-195.
- Yang, H., Establishing acceptable limits of residual DNA. PDA J Pharm Sci Technol, 2013. 67(2): p. 155-163.
- General Chapter<1130> Nucleic Acid-Based Techniques—Approaches For Detecting Trace Nucleic Acids (Residual DNA Testing). United States Pharmacopeia and National Formulary (USP43-NF38).
- World Health Organization. Recommendations for the evaluation of animal cell cultures as substrates for the manufacture of biological medicinal products and for the characterization of cell banks. Replacement of TRS 878, Annex 1. Annex 3. In WHO Expert Committee on Biological Standardization. Sixty-first report. WHO Technical Report Series, No. 978. Geneva: World Health Organization. 2013.
- Guidance for Industry: Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications. US Food and Drug Administration, 2010.
- General Chapter<509> Residual DNA Testing. United States Pharmacopeia and National Formulary (USP43-NF38).
- Guidance for Industry: Analytical Procedures And Methods Validation For Drugs And Biologics. US Food and Drug Administration, 2015.
- Atouf, F. and J. Venema, Do standards matter? What is their value? Journal of Pharmaceutical Sciences, 2020. 109(8): p. 2387-2392.
- Zeine, C., et al., The Value of Pharmacopeial Reference Standards. Pharmaceutical Technology, 2021. 45(2): p. 36-42-36-42.
- World Health Organization. Acceptability of cell substrates for production of biologicals: report of a WHO study group [meeting held in Geneva from 18 to 19 November 1986]. 1987.
- World Health Organization. WHO Expert Committee on Biological Standardization: Highlights of the 46th Meeting, October 19961. Weekly Epidemiological Record= Relevé épidémiologique hebdomadaire, 1997. 72(20): p. 141-145.
 About The Authors:
About The Authors:
Shubrata Khedkar is the bioassay lead at USP India’s biologics lab. She is also an approved instructor for the USP education courses, “Bioassay-Design, Development, and Validation” and “Residual DNA Testing”. Reach her on LinkedIn.
 Bruno De Carvalho, Ph.D., is a member of the USP Biologics marketing team where he supports science communication and provides training to USP staff and global distributors. Before USP, he was a field application scientist for qPCR at Thermo Fisher Scientific, supporting various qPCR applications, including residual DNA detection. He received his M.Sc. from SUNY Albany and his Ph.D. in molecular and cellular pharmacology from SUNY Stony Brook. Reach him on LinkedIn.
Bruno De Carvalho, Ph.D., is a member of the USP Biologics marketing team where he supports science communication and provides training to USP staff and global distributors. Before USP, he was a field application scientist for qPCR at Thermo Fisher Scientific, supporting various qPCR applications, including residual DNA detection. He received his M.Sc. from SUNY Albany and his Ph.D. in molecular and cellular pharmacology from SUNY Stony Brook. Reach him on LinkedIn.