SNP Detection by Enzymatic Means

This is the fourth and final installment in a four-part series on SNP detection. Earlier installments in the series include: SNPs Are All the Rage—Part I, More on SNPs, and SNPs Part III—Putting Instruments to Work.
Introduction
Battle of the Bulge
Invader Assay
Base Excision Sequence Scanning (BESS)
Introduction (Back to Top)
So you want to get into SNP detection, but you're not quite ready to turn in your gel stand for DNA chips and readers. Well, you don't have to. Several enzymatic detection systems are available that, like the solid phase systems described in previous articles in this series, are both high throughput and sensitive. And the good news is that you don't need to buy any new equipment.
Two technologies that apply here both rely on the use of structure specific endonucleases to pick out structural features of DNA duplexes, though in quite different ways. In the simpler case, an endonuclease from the bacteriophage T4 picks out bubbles and bulges in heteroduplexes. Based on the work of Richard Cotton and Rima Youil when at the Murdoch Institute (Melbourne, Australia) and Borris Kemper of the University of Cologne, the enzymatic mutation detection EMD assay was developed for high throughput SNP detection by Variagenics (Cambridge, MA), which is using EMD for SNP screening in its pharmcogenomics operation. Amersham Pharmacia Biotech (Piscataway, NJ) now markets products using this technology under the name Passport kits. Good for a quick scan, this technique requires no knowledge of sequence and hence it is particularly useful in detecting previously unknown, de novo mutations or SNPs.
A second structure specific assay, dubbed Invader by its developers at Third Wave Technologies, (Madison WI) uses a proprietary cleavase in conjunction with a set of sequence-specific probes. The probes are designed to produce a branched structure when hybridized to a homologous target fragment (and not when there's a mismatch), which is then recognized and cleaved by the enzyme. Third Wave offers a variety of detection strategies for analyzing the reaction products, and while not applicable for SNP discovery, Invader has features that make it particularly well suited for high throughput screening.
A third enzymatic detection assay, useful for both mutation scanning and screening, is the base excision sequence scanning system, offered by Epicentre (Madison, WI). With this system, sequence-like fragment patterns are generated from DNA duplexes with modified bases (Ts and Gs) by various base excision schemes.
Battle of the Bulge (Back to Top)
The enzyme mutation detection method revolves around T4 endonuclease VII, one of a number of endonucleases that recognizes structure rather than sequence. Called resolvase in some circles, so-called because it resolves a structural intermediate of recombination (aka Holliday structures), it is unique among enzymes of this type in that it recognizes a variety of DNA substrates—cruciforms (classic Holliday structures), branched DNAs, single-strand overhangs, single base mismatches forming bubbles, or insertions and deletions, causing bulges. Researchers interested in mutation detection seized upon endo VII once it was discovered that it reacts with all kinds of mismatches in double-stranded DNA. Heteroduplexes formed between wild type and mutant DNAs are substrates for endo VII, which, as it cleaves on both strands within a couple of bases of the target, creates fragments that can be analyzed by conventional gel electrophoresis.
The assay typically works by forming heteroduplexes between PCR-amplified wild type and putative mutant fragments, one or both of which can be labeled with an isotopic or fluorescent tag. Once formed, the heteroduplexes are subjected to T4 endonuclease digestion, and the resulting fragments separated by gel electrophoresis. Ordinary one-dimensional gels can be used, but the analysis can be put into higher gear using automated equipment, such as automatic sequencing gels or capillary electrophoesis.
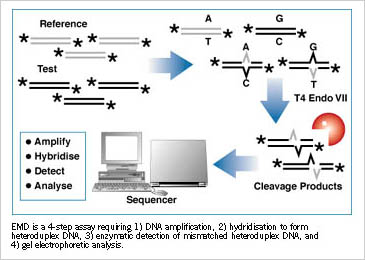
EMD has been tested on various gene systems with all types of DNA including genomic, cDNA, and plasmid preparations. In an early report using EMD to analyze known mutations in beta-globin gene, 81 out of 81 different mutations were detected under a single reaction condition. More recently, in a blind study of unknown p53 mutations in breast cancer cells, 100% of the mutations was detected with EMD. Other genetic systems tested with EMD include CFTR, fibrillin, Rh, BRAC1, and MSMH/MHL1.

EMD has some notable features. Unlike some mutation techniques, for example, the EMD method uses a single, four-step protocol to identify point mutations, deletions, and insertions for all DNA fragments. In addition, DNA fragments several kilobase in size can be assayed directly from PCR reactions. And finally, since the fragments are separated on gels, you can tell not only that a mutation has occurred, but also its approximate location, which facilitates sequencing analysis for confirmation, since only / 30 base pairs around the site need to be analyzed.
Invader Assay (Back to Top)
Third Wave Technologies Invader Assay creates a different kind of substrate, a so-called flap, for a particular structure-specific enzyme, called cleavase. The flap comes about through the hybridization of two, barely overlapping probes to a target. When a complete match is obtained, one probe (the invader probe) invades the second by virtue of a one-base pair overlap, causing the second (the signal probe) to be partially displaced from the hybrid. The structure at the juncture between the flap and the partially invaded duplex is recognized by cleavase, which snips off the displaced part of the probe. Any number of methods, including FRET can detect the released fragment.
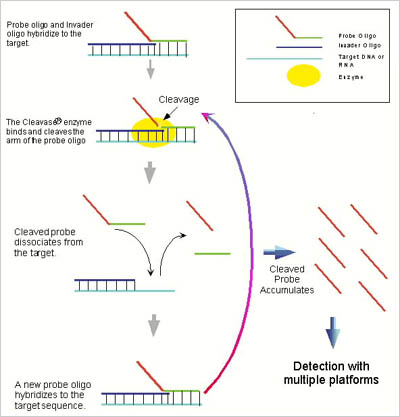
Invader's developers incorporated a number of features into the assay that should make it attractive for high throughput screening studies. By running the reaction close to the melting temperature of the hybrid, a catalytic process is created—the probes continually turn over, allowing for multiple cycles of hybridization and cleavage on each target molecule. This increases the sensitivity to the point that amplification of the target prior to running the assay is generally not necessary. NO PCR! Because of its exquisite sequence specificity (caused by the independent hybridization of two probes) and sensitivity (down to subattomolar concentrations), Invader can pick out mutants in low concentrations in mixtures, 0.1% or 1/1000. And finally, the assay is easily adapted to automation, for example, using a microplate-based FRET format.
Invader assays have been developed by scientists at Third Wave and their collaborators for mutation detection of such clinically important molecules as ApoE, hepatitis B virus, and factor V Leiden. In addition, Endogen (Woburn. MA) has produced a number of Invader assay kits, called Xplore mRNA Assay Kits, for detecting and quantitating mRNA levels—same technology, just with an (unamplified) RNA target rather than DNA. An ever-expanding array of kits is available for detecting some cytokines, both human and mouse, as well as actin, tumor necrosis factor.
Third Wave has already designed dozens of Invader assays for different SNPs, and envisions doing thousands more, all of which (derived from public domain information, anyway) will eventually be posted on a shopping list on its web site. In addition, the company has perfected its design process to the point that it can provide "made to order" Invader assays for your favorite SNP with a couple of day turnaround.
Base Excision Sequence Scanning (BESS) (Back to Top)
Epicentre Technologies (Madison, WI) has combined elements of the two basic sequencing strategies to come up with a non-sequencing approach to genotyping and mutation scanning. Base modification and excision are the centerpieces of its BESS-T and BESS-G Base Reader Kits. With BESS-T and BESS-G, T and G positions can be determined, covering together 100% of mutations. This enzymatic method does not rely on heteroduplex formation or specialized gel conditions, and can be run on ordinary gels or automated DNA sequencing gels.
This three-step process begins by PCR amplifying the region of interest in the presence of limiting dUTP and labeled primers (fluorescent or radioactive). For T-scanning, PCR products are treated with uracil N-glycosylase (UNG), which removes the uracil base from the duplex, producing abasic sites where dUTP was incorporation, and endonuclease IV, which cleaves the fragment at those sites, creating a set of T fragments. Due to a quirk of nature, T-scanning alone, while in theory should only reveal a fraction of mutations, in fact will detect 97% of known human mutations—that is to say, nearly all recorded human mutations involve the loss or gain of a T.
For G-scanning, the same approach is taken, except that prior to excision, the base is chemically modified by a brief light reaction. With both the T and G reaction, strands can be analyzed separately with radioactive labels, or by using different fluorochrome-labeled primers, both strands can be run in a single reaction, which raises the coverage by this technique.
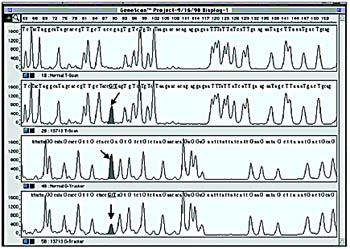
BESS can be easily scaled up by amplifying samples in a 96-well or 384-well plate, for example, or using the automated injection capability on capillary instruments to run multiple samples. As it will pick up both known and unknown SNPs; BESS can be used for either discovery or scoring. And it will pick up a SNP occurring anywhere in the amplified template, which allows the simultaneous discovery of new SNPs and scoring of known SNPs.
A recent report details the use of BESS to characterize genetic variation in 22 isolates of St. Louis encephalitis virus. In addition, Epicentre has posted on its web site BESS based analyses of hepatitis C variants, the human prion gene, and Mycobacterium isolates.
For more information on EMD: Carina Schmidt, Amersham Pharmacia Biotech, SE-751 84, Uppsala, Sweden. Tel: 46 18-16-5744. Fax: 46 18-16-6404. or Fred Ledley, Variagenics Inc., 60 Hampshire Street, Cambridge, MA 02139. Tel: 617-588-5300. Fax: 617-588-5399. Email: fledley@variagenics.com.
For more information on Invader: Sheldon Clark, Director of Marketing, Third Wave Technologies, 502 South Rosa Rd., Madison, WI 53719. Tel: 608-273-8933. Fax: 608-273-6989. Email: sclark@twt.com.
For more information on BESS: Gary Dahl, Epicentre Technologies, 1402 Emil Street, Madison, WI 53713. Tel: 800-284-8474.
By Laura DeFrancesco
