RNA Quantitation -- Down and Dirty
Sub-Attomolar Accuracy
Chromagen's High Performance Signal Amplification
PE Biosystems' Xpress-Screen mRNA Detection Technology
Bayer's QuantiGene mRNA Kits
Sub-Attomolar Accuracy (Back to Top)
For those interested in quantitating a single or small number of RNA species, the high-density expression arrays are not only overkill—spewing out information on thousands of sequences when all you want is one or two—but also they provide only relative quantitation in the form of comparison (ratios) of two samples. While this number may change to four or five as more fluorescent tags become available, still arrays were not designed for experiments with lots of samples, as in drug screening or dose response curves.
The technology profiled here provides sensitivity never even dreamed of (in zeptomolar range, which is 10–21 molar) as well as absolute quantitation of specific RNA species and the capacity to measure in parallel numerous samples. They all work by amplifying the signal rather than the target, freeing you of PCR and the difficulty it presents when dealing with quantitation. Instead, specific probes pull the RNA species of interest out of a lysate, and detectors that combine enzymatic amplification with luminescence generate signal proportional to the signal.
Chromagen's High Performance Signal Amplification (Back to Top)
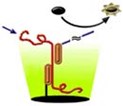
Chromagen's (San Diego) High Performance Signal Amplification System (HPSA) combines solid phase sampling chemistries with a family of fluorogenic dyes, the StarBright dyes, and sensitive fluorescent detection instrumentation for measuring RNA down to the attomolar range. The company has made the technology available as kits for measuring individual mRNA species. Currently 96-well microwell kits are available for measuring TNF-a, IL-6, b-actin, IL-2, and IL-1b, and can be purchased in the U.S. from Oncogene Research Products. In addition, Chromagen offers assay development, screening services, and has configured its tests for high throughput screening (HTS) customers in drug discovery, wishing to measure a particular mRNA or pharmaceutical target.
Chromagen's High Performance Signal Amplification
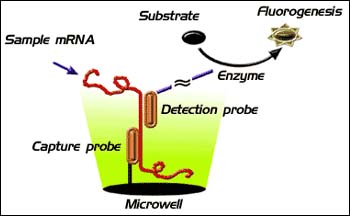
The HPSA direct gene expression assays work by capturing the target mRNA out of a cell lysate using a target-specific capture probe conjugated to the plastic surface of microwell plates. Once captured, the RNA is reacted with biotinylated detection probes that specifically recognize the nucleotide sequence of the target mRNA. The biotinylated probes are then reacted with enzyme-linked (alkaline phospatase or b-galactosidase) StarBright fluorogenic substrates. The amplification afforded by the enzyme coupled with the sensitivity and dynamic range of the StarBright dyes obviates the need for purification or target amplification.
Case Study
Scientist from Chromagen have reported on an assay for measuring adipocyte fatty acid binding protein 2 (aP2) mRNA, a marker for differentiation of preadipocytes into adipocytes, which is transcriptionally activated by the adipocyte specific nuclear hormone receptor PPAR-y. In collaboration with scientists at Merck Research Laboratories, who are evaluating PPAR-y agonists for treating or preventing adipose tissue disorders (insulin resistance, diabetes, and dyslipidemia), Chromagen scientists screened PPAR-y agonists in the mouse cell line 3T3-L1, a commonly used in vitro model of adipogenesis.
HPSA measurements of aP2 mRNA levels in 3T3-Ll cells treated with dexamethasone and insulin in the presence or absence of Merck PPAR-y agonists showed a peak in aP2 mRNA 3 days after treatment, which was confirmed by Northern blot analysis. In addition, determinations of the effective concentration (EC50) using HPSA correlated with results obtained by conventional slot blot hybridization.
For more information: David Blair, Manager, Product Transfer, Chromagen, 10441 Roselle St., San Diego, CA 92121-1503. Tel: 619-558-1456. Email: dblair@chromagen.com.
PE Biosystems' Xpress-Screen mRNA Detection Technology (Back to Top)
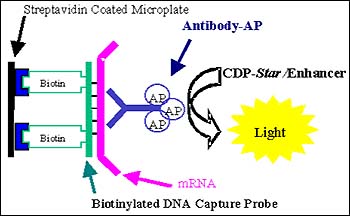
Xpress-Screen technology is offered through the Advanced Discovery Sciences group at PE Biosystems (Foster City, CA). Using custom-designed biotinylated capture probes, RNA is captured in solution from lysates, after which the resulting hybrids are pulled out of solution using streptavidin-coated microwell plates. A proprietary antibody containing multiple alkaline phosphatase conjugates, which binds specifically to DNA/RNA hybrids, is added to the well, followed by a chemiluminescent substrate with enhancer.
PE Biosystems' Xpress-Screen mRNA detection technology.
This combination of reagents provides multiple levels of amplification, resulting in sensitivity down to 0.0017 attomoles of mRNA, which corresponds to 1-2 messages/cell, putting Xpress-Screen among the most sensitive detection available. The amplification levels intrinsic to this method include:
- multiple (2–3) alkaline phosphatase molecules conjugated to each antibody,
- the binding of many antibodies along the length of the DNA/RNA hybrid, and
- processive (continual) cleavage of the chemiluminescent 1,2 dioxetane substrate, which produces a steady glow signal.
Xpress-Screen can be performed on an ordinary luminomter; since glow kinetics are achieved, no injectors are required. The ability to detect as low as 2–3 fold induction means even rare genes can be monitored using Xpress-Screen
Case Study
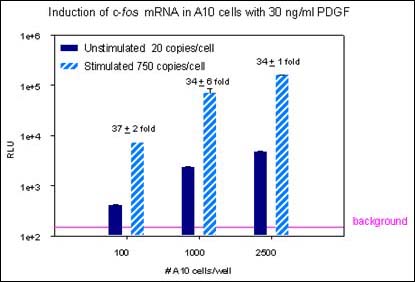
Xpress-Screen technology permits accurate measurement of mRNA levels. The data shown in the bar graph above show reproducibility in a high throughput screen, with a typical co-efficient of variance (CV) below 10%. At even very low cell number (100 cells/well) the fold induction is consistent.
For more information: Dina Martin, Associate Product Manger, Tropix, 47 Wiggins Ave., Befford, MA 01730. Tel: 781-280-5635. Email: martindm@tropix.com.
Bayer's QuantiGene mRNA Kits (Back to Top)
Bayer Corp. (Pittsburgh) offers kits that can be used to quantitate any mRNA species directly from cells with high levels of sensitivity. Using the company's branched DNA (bDNA) technology for signal amplification, in conjunction with a chemilumigenic substrate, it is possible to detect mRNA in the zeptomolar range. And this can be achieved using an ordinary luminometer—the bDNA amplifier does all the heavy lifting.
Working in a microplate format, target mRNA is captured in microwells directly from crude cell lysates using a set of user-synthesized oligonucleotide probes consisting of two probe types: Capture Extenders and Label Extenders. These probes contain sequences complementary to the target mRNA sequence and to sequences common to assay components; in the case of Capture Extenders, complementary to the oligonucleotide coated on the microplate; in the case of Label Extenders, to sequences complementary to the bDNA amplifier. Bayer provides ProbeDesigner software for designing target-specific probe sets from the commonly used GenBank and FASTA databases.
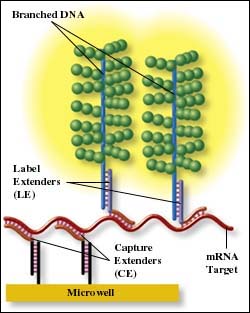
Target mRNA from lysates that is captured in microplate wells is amplified by hybridization with the bDNA amplifier and Label Probes: oligonucleotides complementary to the amplifier that are conjugated to alkaline phosphatase. Each bDNA amplifier can bind many alkaline phosphatase conjugates, thus amplifying the light output signal in the presence of the luminogenic substrate dioxetane. This reaction produces a sustained glow luminescence, so injectors aren't required, an ordinary microplate luminometer will do nicely.
QuantiGene kits come in two varieties: the QuantiGene Expression Kit contains a single strip-well plate with reagents for 96 determinations; the QuantiGene High Volume Kit provides 50 solid plates with reagents for 4800 assays and is adaptable to automated formats for compound screening.
For more information: Stephen H. Chamberlain, Product Manager, Bayer Corp. 4560 Horton St. W810, Emeryville, CA 94608-2916. Tel: 510-923-3079. Email: stephen.chamberlain.b@bayer.com.
By Laura DeFrancesco
