PromoterInspector from Genomatix pulls promoters out of the hat
Applying the company's PromoterInspector tool to the first published sequences from the human genome project (chromosomes 21 and 22), company scientists predicted 50% of the previously characterized polymerase II promoters from "anonymous" DNA sequences. (The majority of all promoters are of pol II type.) In addition, the company believes that among the remaining predicted promoters will be a number of previously unknown promoters.
"We have been eagerly awaiting the complete genome data," states Thomas Werner, founder and CEO of Genomatix. "The tools to annotate promoters for the full human genome have been in place since January 2000. We will be able to finish this project by July 2000."
Werner went on, "In addition, our approach is not to stop just with the annotation. We have also developed the technology to carry out functional analysis of the promoters that will help to reveal the underlying molecular mechanisms for the regulatory pathways involved in the regulation of genes and gene networks."
PromoterInspector
Predicting polymerase II promoter regions is a bioinformatics challenge that up until now has been largely unmet. Polymerase II promoters can be quite different in their individual organization but are recognized in a cell because of a common genomic context. The functional organization of promoters consists of individual transcription factor (TF) binding sites, which are often combined in the form of functional promoter modules. Promoter modules are composed of two or more TF-sites in a defined distance range, which allow synergistic or antagonistic effects.
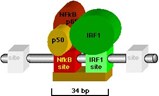
The module below, for example, confers inducibility by tumor necrosis factor alpha (TNF-alpha) and g-Interferon (g-IFN) to several promoters of the MHC/HLA class I genes as well as to b-2 microglobulin and b-interferon genes.

Because of the complex nature of promoters, they cannot be detected reliably by alignment procedures, and general statistical recognition of promoters is hampered by a plethora of false positive matches. However, a promoter, which can be formally seen as a big multi-functional module, can be specifically detected by individual models similar to promoter modules.
PromoterInspector is able to predict promoter regions with high specificity in large genomic sequences, based on context specific features that are extracted from training sequences by a heuristic-free approach, not based on alignment algorithms.
In a recent study of genes induced by TNF and IFN, for example, Genomatrix's model detected the TNF and IFN responsive promoters of HLA-A, B and b-2-microglobulin, though there is no overall sequence similarity between HLA and b-2 microglobulin. In addition, the model discriminated the functionally similar HLA-A and HLA-B sequences from the HLA-C sequences, which do not contain the TNF responsive module, despite high sequence similarity of the HLA-C promoters with HLA-B promoters.
A description of the method used for these promoter prediction is published in a recent issue of the Journal of Molecular Biology (Scherf, M., Klingenhoff, A., Werner, T. "Highly Specific Localization of Promoter Regions in Large Genomic Sequences by PromoterInspector: A Novel Context Analysis Approach," Journal of Molecular Biology 297 (3), 599-606, 2000).
For more information: Genomatix Software GmbH, Karlstraße 55 D-80333 München, Germany. Tel: +49-89-5490839-0. Fax: +49-89-5490839-9. Email: genomatix@gsf.de.
