New Approaches to Cloning: Part I
In the weeks to come, the latest crop of new cloning technologies will be put forth by some of the folks that have brought them to market. Among these are methods that can save you oodles of time with tedious sub-cloning, can relieve you of the necessity of maintaining a freezer full of restriction enzymes, and can deliver a protein to even the most recalcitrant cell line.
First up—transposon-mediated cloning. Jim Pease, product marketing manager at Epicentre Technologies (Madison, WI) provides this description of this novel approach to cloning.
What Is Transposomics?
Transposomics: A Historical Perspective
Transposomics for cDNA and Genomic DNA Sequencing
Transposomics for In Vivo DNA Transposition into the Genome of Living Cells
Future of Transposomics
References
What Is Transposomics? (Back to Top)
Within the past decade, important new techniques have been developed, such as PCR and automated DNA sequence analysis, that have significantly impacted experimental design and expanded our ability to generate data quickly and cheaply. With the recent development of high efficiency DNA transposition systems, a new field of Transposomics is emerging that will further facilitate research in such diverse endeavors as DNA sequencing, functional genomics, and proteomics.
Transposomics constitutes the various in vitro and in vivo applications of transposon-based tools. Transposomics exploits the ability of transposons to be inserted into any target DNA. A simple, high-efficiency transposition system will aid those desiring to sequence cDNA and genomic DNA, map cloned or chromosomal DNA, create gene "knockouts," or to insert any desired DNA sequence into any target DNA in vitro or in vivo.
Transposomics: A Historical Perspective (Back to Top)
Transposons are mobile DNA sequences found in the genomes of prokaryotes and eukaryotes. Naturally occurring transposons are composed of a DNA sequence contained between specific transposase recognition sequences (the transposon Outer Ends, or OEs). The transposase enzyme catalyzes the transposition process. Frequently, the transposase gene is contained within the transposon and must be expressed for transposition to occur. In some systems, host cell proteins also play a role in the transposition process.
Transposition systems have been exploited for chromosome mapping, to provide physical landmarks within chromosomes, and for inserting selectable markers into organisms. However, several inherent problems associated with transposition systems have limited their widespread use. For example, wild-type transposition occurs naturally at a very low frequency. In addition, some transposition reactions require propagation and mating of bacteria strains, expression of the transposase, and involvement of host cell factors. Finally, many systems exhibit transposition "hot" spots and "cold" spots within the target DNA.
In 1998, Goryshin and Reznikoff (ref. 1) developed a high-efficiency transposition system that can be used for both in vitro and in vivo transposition reactions. This simple yet elegant Tn5 transposition system circumvents many of the previously mentioned problems.
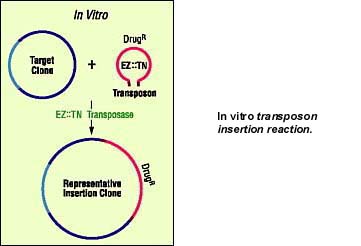
The advantages of the Tn5 transposition system include:
- Tn5 transposase is a small, single subunit enzyme that has been cloned and purified to high activity. A cloned transposase means that a functional transposition system can be reconstituted in vitro.
- Tn5 transposase carries out transposition without the need for host cell factors.
- Tn5 transposition proceeds by a simple "cut and paste" process that appears to insert transposons with nearly perfect randomness into the target DNA.
- Tn5 transposase will transpose any DNA sequence contained between its short 19-basepair OEs.
In addition, mutations have now been engineered into both the Tn5 transposase gene and its 19 basepair OEs that result in a hyperactive system with a transposition efficiency 1000-fold greater than wild-type. This important breakthrough provides the opportunity to develop reliable, simple and efficient transposon-based tools for a wide variety of in vitro and in vivo applications.
Transposomics for cDNA and Genomic DNA Sequencing (Back to Top)
The power of Transposomics as applied to DNA sequencing has been demonstrated by Jendrisak, et al (ref. 2). Current strategies for sequencing large DNA (>2 kb) cloned into plasmid, cosmid, or BAC vectors require reducing the cloned DNA to manageable sizes (e.g. 0.5–2 kb) by creating deletions with exonuclease III or mung bean nuclease or by random subcloning of DNA fragments. However, these approaches are tedious and labor intensive, thus reducing the overall efficiency of the sequencing project. "Primer walking" (in which the initial sequence obtained is used to design primers for obtaining adjacent sequence data) provides an alternative approach, but with a high rate of primer failure, it can quickly become a costly and time-consuming procedure.
Using a transposition system based on Tn5 (currently marketed under the tradename EZ::TN by Epicentre Technologies), Jendrisak and coworkers inserted a transposon containing sequencing priming sites and a kanamycin resistance marker randomly into a plasmid clone in vitro. Kanamycin-resistant transformants (transposition clones) were selected and subsequently sequenced bi-directionally from the priming sites contained on the inserted transposon. Due to the high transposition efficiency of the system, a single in vitro transposon insertion reaction generated sufficient clones to sequence the entire insert.
Using a somewhat different approach, the same group sequenced a 6.3 kb DNA that had been cloned between the Tn5 OE sequences contained in a specially constructed plasmid (pPDM). They first created a population of sequence deletions within the insert DNA via an intramolecular transposition reaction in vitro. Deletion clones were screened and subsequently sequenced from priming sites on the vector. Again, a single, simple in vitro transposition reaction generated sufficient deletion clones to sequence the entire insert in less time and with significantly less effort than generating deletions using exonuclease III and mung bean nuclease.
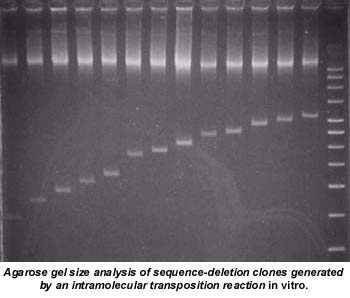
Thus, Transposomics provides a more efficient and cost-effective way to generate templates for sequencing large DNA than exonuclease III/mung bean nuclease digestions, random subcloning, and primer walking. The Transposomics approach to generating DNA sequencing templates should appeal to both high-throughput sequencing facilities as well as smaller sequencing operations.
A Tn7 transposition system developed by Stellwagen and Craig (ref. 3) is available from New England Biolabs (Beverly, MA) under the name Genome Priming System. Like the Tn5 system, it sports a vastly simplified tranposition system that inserts randomly and at high frequencies.
Transposomics for In Vivo DNA Transposition into the Genome of Living Cells (Back to Top)
Although Transposomes (the complex formed between transposase and transposon) are normally formed transiently during the transposition process, a stable Transposome can be formed and isolated in vitro in the absence of magnesium. It has recently been demonstrated that Transposomes can be electroporated directly into living bacteria (ref. 3) and Saccharomyces (ref. 4), and subsequently it will carry out stable integration of DNA into the genome of these organisms. Presumably, once inside a cell, the Transposome is activated by Mg2+ within the cell and then randomly inserts its transposon into the host cell's chromosomal or extra-chromosomal DNA.
In one demonstration of in vivo transposition, a Transposome containing the kanamycin MX4 antibiotic resistance marker was electroporated into Saccharomyces. Using Geneticin (G418) selection, transformants were selected and analyzed. Successful integration into the cell's genome was demonstrated by PCR. No transformants were obtained by electroporation of the kanamycin MX4 transposon alone. The genomic DNA of the transformed yeast has since been cloned into a cosmid vector. Transposon integration sites, rescued as kanamycin-resistant cosmid clones, can now be sequenced or mapped to reveal more about the nature and precise locations of the transposon insertion sites.
Results from this early work indicate that high efficiency in vivo transposition may become the method of choice for introducing DNA into some cells.
Future of Transposomics (Back to Top)
The development of the hyperactive Tn5 transposition system opens up a myriad of opportunities to utilize Transposomics for both in vitro and in vivo applications. Insertion of transposons in vitro will be used for DNA sequencing, creating gene "knockouts" mapping cosmid or BAC clones or inserting any DNA sequence of interest into any DNA target. Transposon-mediated sequence deletion reactions in vitro will be used to generate templates for DNA sequencing, creating truncated genes for structure-function studies, epitope mapping, and for identifying the exact start and stop sites of cloned genes. In addition, because theTn5 transposase will transpose any DNA sequence contained between its OEs, custom transposons can be produced for user-specific applications.
One of the more intriguing applications of Transposomics is the stable insertion of DNA sequences into the genomes of living cells in vivo by direct electroporation of Transposomes. It has been demonstrated that selectable resistance markers can be integrated into and expressed from the genome of bacteria and yeast. An obvious extension of this work is to create custom Transposomes, containing any DNA sequence of interest, for integration into the genome of cultured cells of higher organisms (e.g. Homo sapiens).
- Goryshin, I. and Reznikoff, W. "Tn5 in vitro transposition", J. Biol. Chem., 273: 7367-7374 (1998)
- Jendrisak, J. et. al., "High efficiency in vitro transposition for DNA sequencing projects", 10th International Genome Sequencing and Analysis Conference, Miami Beach, FL (1998)
- Goryshin, I. and Reznikoff, W., personal communication
- Hoffman, L.M., et. al. "In vivo transposition of transposon/transposase complexes into the genome of Saccharomyces", XIX International Conference on Yeast and Molecular Biology, Rimini, Italy (1999).
Jim Pease has a Ph.D in Biochemistry, from the University Illinois-Urbana. After working in the industry for such giants as Pharmacia, Boehringer Mannheim and Promega, he came to Epicentre to be a product and marketing manager. Jim knows all aspects of bioresearch product development, having held positions in technical support, production, training and QC.
For more information: Jim Pease, Product Marketing Manager, Epicentre Technolgoies, 1402 Emil St., Madison, WI 53713. Tel: 608-258-3080 or 800-284-8474. Fax: 608-258-3089. Email: jim.pease@epicentre.com.
