Leveraging AI And Data Science For Biologics Characterization
By Simon Letarte, Letarte Scientific Consulting LLC

Biologics complexity presents significant challenges for characterization, quality control, and regulatory approval. Unlike small molecules, biologics are heterogeneous and structurally intricate, requiring advanced analytical techniques for comprehensive characterization. In this context, artificial intelligence (AI) and data science have emerged as powerful tools, augmenting traditional characterization methods and driving innovation throughout the life cycle of biologic drug development.
Artificial intelligence encompasses machine learning (ML), deep learning, natural language processing (NLP), and other computational techniques that enable software to learn from data, identify patterns, and make predictions. Data science refers to the broader discipline of collecting, processing, analyzing, and visualizing data to extract actionable knowledge.
This paper will focus on the three following case studies:
- CQA data sharing
- Peptide map results interpretation
- Automated regulatory authoring
Distilling Raw Data Into CQA Measurements
The characterization of biologics encompasses detailed analysis of their physicochemical properties, including structural integrity, glycosylation patterns, aggregation status, purity, potency, and immunogenicity. Analytical platforms such as mass spectrometry, chromatography, electrophoresis, and bioassays generate complex data sets that require expert interpretation, often through several layers of software. For example, an LC-MS experiment has raw data that is composed of retention time, m/z, and intensity. The raw data tends to be very large and not very informative on its own. It needs to be contextualized and go through a software stack to identify the peptides and proteins that were contained in the sample, as illustrated in Figure 1. Most of the time, we are not interested in the peptides and proteins but in the post-translational modifications (PTMs) on specific amino acids. Therefore, another step of data processing is often necessary.

What data do we share, and how? The raw data should be kept for as long as the corporate policy will allow so it can be re-interrogated if needed in the future. Realistically, the raw data is only useful to the team that performed the analysis because it requires specialized software to read and process. It is the processed data that is of interest to the broader team, and it should be easily accessible to the stakeholders.
Mass spectrometry is unique among analytical techniques because it can tell what is in the sample and how much of it there is. This poses a problem for standardization of the data because the reported attributes may not be defined until after the analysis is complete.
One solution to sharing analytical data is to focus on critical quality attributes (CQAs). In early stages of development, exploratory experiments should be performed to identify the molecule’s liabilities and degradation pathways. Once that is established, one must create analysis templates to record the value of each potential CQA so that every time the analysis is performed, those values are recorded in a structured way. For this to be successful, the team needs to align on a standard vocabulary to name attributes.
Central to this concept, a CQA database is a useful tool, where attributes, criticality scoring, justifications, and recorded values can be visualized for each attribute. One can later search for maximum, minimum, or average value for any lot of product analyzed. This information needs to be available cross-functionally to all stakeholders.
Using AI To Interpret Peptide Map Results
Another interesting area where AI can be applied is in the interpretation of analytical testing results. AI could look at the peptide map results and tell the analyst what the concerning PTMs are as well as their impacts on PK/PD, biological activity, immunogenicity, and safety. This is a level of knowledge that an expert analyst would have in their head, but leveraging AI could make everybody an expert.
General large language models (LLMs) can perform this task to a certain extent but can be unreliable when the residues don’t match the Kabat notation. Most current AI tools do not understand biology and cannot account for whether an amino acid is in an impactful region or not. But this is changing very rapidly, and the tools are much better today than they were a few months ago. There are opportunities to make dedicated AI tools, such as small language models (SMLs) and fine tune them to produce more comprehensive and reliable results for a set of predefined tasks.
Automated Regulatory Authoring
Regulatory authoring tends to be a time-consuming and stressful activity for the analytical scientist. This includes wrapping up the last experiments, finishing the electronic lab notebooks (ELNs), writing up the scientific reports, the multiple rounds of revisions, and, finally, authoring the regulatory document. To streamline this process, one can develop content mapping scripts between data from an ELN to automatically transfer tables and figures into a predefined section of a Word document, as illustrated in Figure 2. Once the notebook is approved, the data remains in a validated state and can be mapped into the regulatory document template.
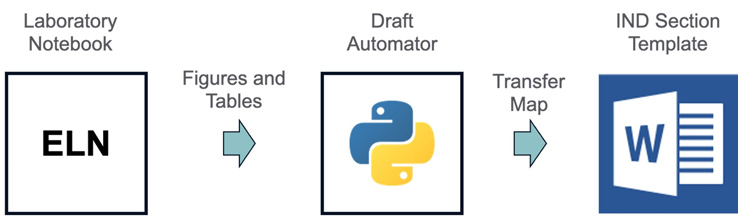
The above approach encourages end-to-end planning of the regulatory writing and can save significant time iterating across the different versions of the reports. Most modern ELNs offer application programming interface (APIs) that can interface with custom scripts to transfer the data into Microsoft Word documents.
A Word Of Caution
Although AI can write entire documents, we need to keep in mind how the activities of writing and thinking are interconnected. It is well documented that writing things down helps with remembering them and creates connections in the brain.1, 2
As an illustration of this process, I was recently reviewing a report one of my team members wrote. We had found a trace-level impurity but were unsure of its identity. The deadline for the report was approaching, so we began drafting. As I read the draft and rewrote some sections, something clicked. It clicked because I was reading the entire body of evidence at once and mentally connecting loose ends. My attention was drawn where the text was imperfect, perhaps a sign of gaps in the story. I started thinking and rearranging paragraphs. Eventually, I noticed minor peaks from another assay that were explaining our unknown trace impurity.
The process of writing and revising the document directly led to this important insight. I spent more time where the text was imperfect and unclear, which indicated to me a lack of understanding and deficiency in the data. If the text had been written by an AI, all proper, grammatically correct, and boring, I might have missed it.
Challenges And Future Directions
While the promise of AI and data science in biologics characterization is profound and goes far beyond the few case studies presented here, several obstacles remain:
- Data quality and standardization: The utility of AI depends on robust, high-quality training data, which is often lacking due to proprietary constraints and variability in experimental practices. As with anything, garbage in, garbage out.
- Model interpretability: Regulatory agencies and stakeholders require transparent, interpretable models. The “black box” nature of deep learning can limit adoption. We need more deterministic and fewer probabilistic models.
- Integration with legacy systems: Many laboratories operate with heterogeneous equipment and software, complicating data integration and workflow automation. A lot of effort is needed at the interfaces to ensure the connections are made correctly.
Future advances may include federated learning for cross-institutional model development, use of specialized SMLs, explainable AI for regulatory acceptance, and the integration of AI with real-time analytics for adaptive process control.
Conclusion
Today is a great time to enroll AI to give a productivity boost to your team. As resources are stretched thin across organizations, these tools allow scientists to focus their time on value-added work. AI can impart advanced knowledge to junior scientists so they can perform tasks that only the most senior team members could do only a few years ago. This shift has the potential to lower the cost of developing safe, effective medicines and increase profitability of biotech companies.
References:
- Hillesund, T., Scholarly reading (and writing) and the power of impact factors: a study of distributed cognition and intellectual habits, Frontiers in Psychology, Vol 14, 2023.
- Menary, R., Writing as Thinking, J. Lang. Sci., Vol 29, No. 5, 2007.
About The Author:
 Simon Letarte, Ph.D., is an analytical chemist and principal consultant at Letarte Scientific Consulting LLC. He has more than 15 years of experience characterizing proteins and peptides using proteomics and mass spectrometry. Previously, he was a director of structural characterization at Gilead Sciences. Before that, he worked as a principal scientist at Merck and, before that, Pfizer. Other previous roles include senior research scientist roles at Dendreon and the Institute for Systems Biology. Connect with him on LinkedIn.
Simon Letarte, Ph.D., is an analytical chemist and principal consultant at Letarte Scientific Consulting LLC. He has more than 15 years of experience characterizing proteins and peptides using proteomics and mass spectrometry. Previously, he was a director of structural characterization at Gilead Sciences. Before that, he worked as a principal scientist at Merck and, before that, Pfizer. Other previous roles include senior research scientist roles at Dendreon and the Institute for Systems Biology. Connect with him on LinkedIn.
