Human Genome Sciences and Compugen to Create Map of All Human Expressed Sequences
The analysis of the gene data will include more than 2.5 million mRNAs that HGSI has isolated from all human organs and tissues from 800 individuals—a collection the company believes represents more than 95% of all human expressed genes. Information from public databases—including the entire nucleotide sequence of chromosome 22, more than one billion nucleotides of human genome sequence, and sequences of more than one million messenger RNAs, as well as any additional sequences released over the next year—will be folded into the analysis.
cSNPs Mapping
The HGSI/Compugen collaboration should generate a large collection of single nucleotide polymorphisms in expressed genes. As the HGSI collection includes gene samples obtained from more than 800 unrelated individuals, and there are an estimated four polymorphisms per individual messenger RNA of any two people, there could be more than 500,000 cSNPs in its gene collection of 2.5 million messenger RNAs. New computation methods for comparing nucleotide differences in the messenger RNAs obtained from 800 different individuals will identify such variants.
Gene Organization and Splice Variants
Since the Compugen LEADS technology allows direct comparison of a mRNA sequence with the corresponding chromosomal sequence, the structure of the gene and the positions of exon-intron boundaries will be revealed. HGSI and Compugen scientists will analyze the results of such comparisons on a gene by gene basis, resulting in a comprehensive analysis of human gene organization.
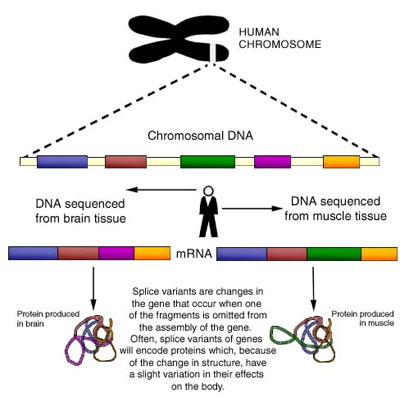
The Compugen LEADS technology is also designed to detect variation in splicing of individual genes. It's known that the final structure of a messenger RNA may vary between organs and tissues, depending on how the genes were spliced together to create a messenger RNA. Spliced variants may produce proteins with substantially different functions, and could account for substantial differences in the function of individual cells, organs and tissues. The Compugen LEADS technology has been specifically designed to detect splicing variants.
Steve Ruben, vice president of research of HGSI, said: "… The biochemical and biological function of a gene and of a protein can vary from organ to organ. In the most extreme use the two spliced variants may have opposing biological functions. For example, we have discovered several spliced forms of the gene messenger RNA that produces human growth hormone. Most of these spliced variants produce proteins that promote growth. However, at least two of the spliced variants we discovered have the opposite effect, i.e., produce proteins that inhibit growth. Having extensive knowledge of human spliced variants should substantially accelerate our search for new biological human genes and proteins to help treat and cure disease."
Human Genome Sciences's mission is to develop products to predict, prevent, detect, treat, and cure disease based on the discovery and understanding of human and microbial genes. Compugen develops and utilizes advanced computational technologies for genomic and proteomic research. The core technologies of the company result from a multidisciplinary research effort combining advanced mathematics and related disciplines with molecular biology. The company's proprietary products and services are commercialized to other organizations in the form of corporate solutions, such as the LEADS platform, and to individual life scientists worldwide over the company's internet research engine.
For more information: Compugen Inc., R&D and Customer Support Center, 7 Centre Dr., White Oak Building, Suite 7, Jamesburg, NJ 08831. Tel: 609-655-5105. Fax: 609-655-5114.
