Finding the proverbial needle
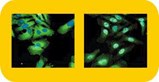
Cellomics' HCS system for drug discovery enables large-scale screening of multiple targets in intact cells, and their cell-based assay kits can also be used at the basic research level.
Contents
Introduction
High Content Screening
HitKits
References
Introduction (Back to Top)
Cellomics, Inc. is a privately held company that specializes in products and services designed to help the pharmaceutical sector achieve faster and more effective drug discovery. The enormity of the challenges faced by drug discovery companies was aptly described by Alan Dove, contributing editor to Nature Biotechnology, who wrote, "Screening for a drug is often likened to searching for a needle in a haystack. In fact, with the application of combinatorial approaches and parallel synthesis to discovery, the sheer scale and diversity of chemical libraries have made identifying drug leads more akin to searching for a needle in many different haystacks"1. Indeed, contemporary drug screening efforts often process up to 100,000 compounds a day against a rapidly expanding universe of drug targets. To meet these challenges, "big pharma" is turning more and more to smaller, specialty biotechnology companies such as Cellomics for help. As described in more detail below, Cellomics' cell-based assays were designed and optimized for use in large-scale screens in which multiple properties of scores of individual cells are characterized via use of the company's ArrayScan II System. However, if you are a basic researcher working on a smaller scale, don't stop reading. The fluorescent probes and immunochemical reagents employed in the cell-based assays are fully compatible for use with standard fluorescence microscope systems, and can be used in basic research applications.
High Content Screening (Back to Top)
Cellomics' unique High Content Screening (HCS) platform exploits advances in cell biology and harnesses the power of fluorescence imaging, bioinformatics, and robotics for the screening of multiple targets within intact cells. Unlike conventional plate readers, the HCS system enables measurements of fluorescently labeled targets within individual cells in each well. This allows researchers to monitor the biological variability of cells in a given assay, as well as to isolate and analyze cell sub-populations within a given microplate well. In the HCS approach, specific cellular constituents are first fluorescently labeled using any of a number of the company's HitKit series of reagent kits, then analyzed using the ArrayScan II System. This system uses automated image acquisition to gather information about the spatial and temporal distribution of fluorescently labeled cellular components of cells grown in microtiter plates. The ArrayScan software then enables automatic image processing for measurement of various effects of new drug candidates in the cell. It quantifies multiple fluorescent signals on or in cells, and converts raw image data into parameters such as subcellular distribution, area, morphology and activity measurements. Cell fields, exposures, thresholds, and focus parameters are automatically set during plate scanning, and "real-time" data access is provided during scanning. Images and data are also stored for later use, enabling "walk away" operation. The ArrayScan II imaging system is currently on the market and according to Cellomics, has already been purchased by several major pharmaceutical companies. In 1998, researchers at Merck Research Laboratories (Rahway, NJ) and collaborators at Cellomics reported the use of this imaging system's forerunner (ArrayScan) for fluorescence-based analysis of NF-kB translocation to the nucleus 2.
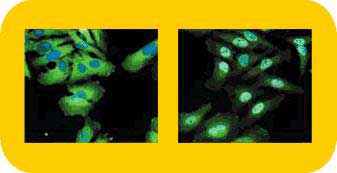
HitKits (Back to Top)
The HitKit series of HCS kits contain proprietary and licensed fluorescent reagents, chemicals, and in some cases, cells. Optimized sample preparation protocols and assay instructions for using the kits in conjunction with the ArrayScan II System are also included. Although HitKit protocols have been validated and optimized for use with the ArrayScan II System, these kits can also be used in conjunction with fluorescence microscopes in basic research applications. Cellomics currently offers fifteen different HitKits, and more are under development. As described in more detail below, this diverse group of kits includes those for determining cell viability, tracking the amounts and subcellular locations of key proteins, and monitoring cell morphology or motility. Studies utilizing more than one HitKit can be readily developed because of the multi-parameter nature of HCS. Thus, researchers can create specificity assays in which two or more proteins or pathways can be simultaneously measured in individual cells.
Wanted: Dead or Alive
The Cell Viability HitKit is used for assaying the number and percentage of live and dead cells on any standard microplate or microscope slide. This kit employs two proprietary fluorescent stains, DeadDyeand VitalDye. DeadDye enters dead (or dying) cells via leaky plasma membranes. It binds to nucleic acids, and can be visualized by its red fluorescence emission. VitalDye is a membrane-permeable live-cell indicator that emits green fluorescence when cleaved by endogenous cellular esterases. Hoechst 33342 is used to label the nuclei of all cells in a population, and is included in each different type of HitKit. This membrane-permeable dye binds nucleic acids and emits blue fluorescence.
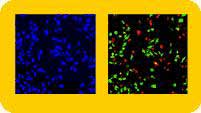
Hitting the Target
Numerous HitKits employ unlabeled primary antibodies specific for the target molecule of interest and a fluorescent secondary antibody conjugate. This immunochemical strategy is used in all of the assays for visualizing and quantifying the nuclear translocation of signal transduction molecules. HitKits for signal transduction studies include those specific for STAT transcription factors (STAT1, STAT2, STAT3 or STAT5), NF-kB, c-jun, ERK (MAPK), p38 MAPK (HOG1) and JNK/SAPK. The Mitotic Index and Neurite Outgrowth HitKits also employ this same basic type of antibody-based labeling tactic. The Mitotic Index HitKit is used to characterize proliferating cells and to identify compounds that inhibit or induce mitotic progression. It employs an anti-histone antibody and fluorescent secondary conjugate to specifically label mitotic cells. The Neurite Outgrowth HitKit enables the specific immunodetection of neuronal cell bodies and neurites in heterogeneous cell populations. The Transferrin Receptor HitKit utilizes a different type of labeling strategy. In this assay, a fluorescent conjugate of human transferrin is used to follow receptor internalization and trafficking.
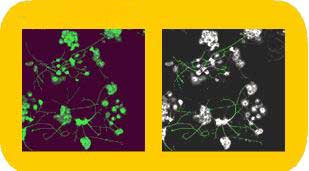
The Multiparameter Apoptosis 1 HitKit allows investigators to characterize three different properties that vary as cells undergo apoptosis: nuclear morphology (condensation/fragmentation), mitochondrial mass/potential, and the cytoskeletal F-actin content. Nuclear morphology is examined by use of Hoechst 33342. Changes in mitochondrial membrane potential are followed by use of MitoTracker Red, a cell-permeant, mitochondrion-selective fluorescent dye developed and patented by Molecular Probes, Inc. (Eugene, OR). Cytoskeletal F-actin content is examined via use of a fluorescent conjugate of phalloidin, an Amanita phalloides mushroom toxin that specifically binds F-actin filaments. The recently launched Cell Spreading HitKit is used to quantify cell spreading by measuring the size of fluorescently labeled cells. Rhodamine-conjugated phalloidin is employed for cell area measurements.
Cellomics also provides contract custom assay development services for drug discovery companies.
References (Back to Top)
1. Dove, A., "Drug screening—beyond the bottleneck," Nature Biotechnology 17:859-863, 1999.
2. Ding, G.J.F. et al., "Characterization and quantitation of NF-kB nuclear translocation induced by interleukin-1 and tumor necrosis Factor-a: Development and use of a high capacity fluorescence cytometric system," JBC 273:28897-28905, 1998.
For more information: Cellomics Inc., 635 William Pitt Way, Pittsburgh, PA 15238. Tel: 412-826-3600. Fax: 412-826-3850. Email: info@cellomics.com.
About the author:
Deborah A. Fitzgerald earned a Ph.D. in biochemistry at the University of Tennessee, then completed her first postdoctoral fellowship in U.T.'s Division of Hematology. After conducting research at St. Jude's Children's Research Hospital (Memphis, TN) for three years on the molecular basis of pediatric leukemias, she became a freelance biotechnology writer. She currently resides in Birmingham, AL, and can be reached at deb@sciwriters.com.(Back to Top)
