DNAZap: Nucleic Acid Degrading Solution
By Timothy W. Abraham and Tiffany J. Smith
Although the polymerase chain reaction (PCR) has revolutionized molecular biology, the benefits of exponential amplification have been accompanied by the necessity of avoiding nucleic acid contamination leading to false-positive results. This is especially true in the fields of diagnostics and forensics. Measures currently taken to avoid cross-over contamination often fall short of this goal. DNAZap (patent pending) provides a quick, convenient, and reliable method for pre-PCR decontamination and can be used to safely decontaminate any surface, including thermocycler blocks and lids, pipettors, gloves, microcentrifuge tubes, tube racks, ice boxes, and work benches.
Introduction
Methods and Results
Summary
About the Authors
Introduction (Back to Top)
PCR is used to detect small amounts of a specific target sequence, providing a powerful method for assaying low abundance nucleic acids and enabling researchers to identify a gene or transcript from a single copy of a specific sequence. PCR has therefore become invaluable in the molecular diagnosis of genetic diseases, identification of bacterial and viral pathogens, and forensic identification.
A problem commonly encountered in PCR is the amplification of contaminating nucleic acids. The contamination of samples and/or reagents renders PCR subject to false-positive results and therefore places a limitation on its use in routine diagnostic applications. False-positive results can arise from carry-over products from previous PCR or cross-contamination between samples or with positive controls. Benchtops, pipettors, gloves, microcentrifuges, iceboxes, tube racks, and labeling pens are common sources of contamination.
While it is prudent for researchers to take preventative measures to avoid nucleic acid contamination, many of the methods currently used are inadequate, cumbersome, and harsh on equipment. For example, pipettors and other surfaces are treated with caustic bleach solutions or exposed to UV light. Experimental reagents may be exposed to UV light or ionizing radiation. DNAZap is a nucleic acid decontamination solution that effectively and efficiently degrades nucleic acids from all types of surfaces almost instantaneously without damaging expensive equipment or compromising PCR or other downstream enzymatic reactions.
Methods and Results (Back to Top)
DNAZap consists of two solutions, each of which alone is inefficient in degrading nucleic acids. Combining the two solutions generates a short-lived, potent, nucleic acid-degrading mixture, quickly degrading high levels of contaminating DNA and RNA. The first solution is applied to the surface to be decontaminated followed by the immediate application of the second solution. A thorough rinse with distilled water washes away any by-products including the degraded nucleic acids.
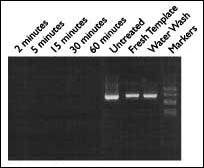
A sensitive PCR experiment demonstrating the effectiveness of DNAZap in rapidly removing contaminating DNA from PCR tubes is shown in Figure 1. Template DNA was dried down in several PCR tubes and then treated with distilled water as a negative control or with DNAZap for different periods of time. PCR carried out in these tubes treated with DNAZap did not yield any detectable products, even in tubes treated with DNAZap for as little as 2 minutes.
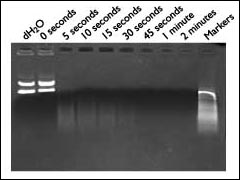
A time course experiment demonstrating the ability of DNAZap to rapidly degrade nucleic acid is shown in Figure 2. Replicates of plasmid DNA were treated with DNAZap, and the reaction was quenched at various time points. Most of the DNA was degraded within the first few seconds, and only traces of DNA remained at 30 seconds. It must be emphasized that the solutions are most effective immediately after mixing. Thus, they cannot be pre-mixed and stored for later use.
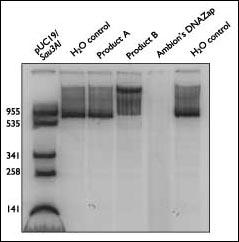
Many products that claim to be decontamination solutions act only as detergents, essentially diluting and spreading the contaminating nucleic acids. In contrast, DNAZap is the only product available that actually degrades nucleic acids. Figure 3 demonstrates this exceptional property of DNAZap. For this experiment, equal amounts of [a-32P]dATP-labeled PCR product were treated with DNAZap, other commercially available "DNA decontamination products," or distilled water. The DNA treated with DNAZap was degraded to single nucleotides, whereas the other solutions left large DNA fragments. Other experiments using a 32P-labeled RNA transcript gave similar results (data not shown).
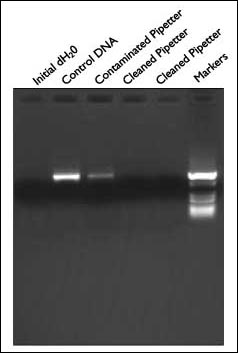
A serious concern among researchers performing PCR is contamination from aerosols created in pipettors. Figure 4 demonstrates the effectiveness of DNAZap in addressing this problem. First, a clean pipettor was intentionally contaminated with template DNA. The contaminated pipettor was used to transfer distilled water to a PCR tube. This pipettor was then taken apart and the plastic barrel and the stainless steel ejector arm were treated with DNAZap and rinsed. Distilled water was subsequently transferred to a PCR tube using the cleaned pipettor. PCR was performed in these tubes without adding any template DNA. Analysis of the PCR products showed that the contamination from the pipettor evidenced in the first tube was completely eliminated by treatment with DNAZap.
Summary (Back to Top)
When accuracy and reliability are essential in performing PCR, the exceptional DNA and RNA degrading properties of DNAZap provide the researcher with a quick and easy method to safely clean contaminated surfaces and prevent sample-to-sample contamination. DNAZap is also useful in avoiding contamination in many other applications including the ligase chain reaction, Q-beta replicase-mediated amplification, strand displacement amplification, the repair chain reaction, self-sustained sequence replication, and nucleic acid sequence-based amplification (NASBA).
About the Authors (Back to Top)
Timothy W. Abraham (Senior Scientist) and Tiffany J. Smith (Product Manager) are employed by Ambion, Inc. (Austin, TX).
For more information: Ambion, Inc., 2130 Woodward St., Suite 200, Austin, TX 78744. Tel: 800-888-8804.
