ProteinChip System Creates Inroads into Phenomics
Contents
Introduction
Why Phenomics?
What is the ProteinChip System?
Bio-Marker Discovery Studies Performed Using Seldi ProteinChip Systems
Case Study 1: Prostate Cancer
Case Study 2: Ovarian Cancer
Case Study 3: Alzheimer's Disease
Seldi Protein Biology System II
References
Introduction (Back to Top)
Genomics and proteomics move over. There's a new "-omic" on the horizon: phenomics—defined as a study of the molecular changes that give rise to particular phenotypes, something every drug discovery and pharmaceutical company wishes to know. And now there's a new tool for bringing phenotypic data into focus. Ciphergen Biosystems Inc. (Palo Alto, CA) has developed the ProteinChip System that can quickly and sensitively profile the spectrum of proteins in very small samples. By combining "on-chip" affinity purification and secondary chemical or enzymatic processing with mass spectroscopy, this tool provides specificity, sensitivity, and speed unobtainable with any other single system.
"Much as PCR and DNA chips were enabling tools for genomics, we think the ProteinChip System is a revolutionary alternative to existing methods of protein biology, said William E. Rich, president and CEO of Ciphergen. "In as little as a day, researchers can discover, identify, and characterize disease-related proteins from biological fluids and tissue, and proceed immediately to analyze protein function—all on the chip."
Why Phenomics? (Back to Top)
A complete understanding of normal and disease states can never be obtained from genomics research alone. While genotyping may identify the genes involved in disease or the predisposition to disease, and even their expression level, it cannot provide information on protein expression, protein-protein interactions, post-translational modifications, and processing of proteins. Current approaches to these questions require stringing together several technically difficult methodologies—two-dimensional gel electrophoresis and mass spectroscopy, most commonly—and can be cumbersome and difficult to scale-up.
What is the ProteinChip System? (Back to Top)
At the core of Ciphergen's ProteinChip System lies Surface-Enhanced Laser Desorption/Ionization (Seldi), the brainchild of T. William Hutchens, board member of Ciphergen, and his colleague T.T. Yip, now director of research at Ciphergen, while at Baylor College of Medicine (ref. 1). This technology enables fully integrated "on-chip" processing of proteins, including protein capture, protein structure and function evaluation, and molecular detection.
The ProteinChip Array contains hundreds of multiple, addressable locations of chemically or biologically defined protein binding surfaces arranged as wells of 1mm diameter or smaller. The arrays contain various kinds of functional groups—from antibodies and receptors to chromatographic surfaces—to capture proteins. Minute quantities of samples such as serum, urine, or tissue extracts, can be incubated directly on the chip, without having to purify the proteins first. After the chip has had a chance to bind molecules in the sample, and unbound material is washed away, it is treated with short bursts of laser light, releasing and ionizing the proteins, which are accelerated to a detector. The time-of-flight required for proteins to reach the detector is calculated to determine the molecular weight of each released protein. The result is graphically displayed as a protein profile of all captured proteins from a particular sample. The ability to use multiple array surfaces amplifies the ability to rapidly define complex protein compositions by simultaneously investigating a variety of purification conditions.

Bio-Marker Discovery Studies Performed Using Seldi ProteinChip Systems (Back to Top)
The search for biomarkers in cancer cells, Alzheimer's disease patients, and pathogenic bacteria has already benefited from the ProteinChip platform. Profiling differentially expressed proteins has identified proteins in prostate, bladder, and ovarian cancer cells that might provide diagnostic markers for early detection and disease progression. The ProteinChip Array was also used to develop an assay for different amyloid beta peptides, potential diagnostic markers for Alzheimer's disease onset. Efficacy of drug treatment and identification of protein markers has also been demonstrated with the pathogenic microorganisms, M. Tuberculosis, H. influenzae, and mycoplasma. Protein markers from these microorganisms, identified by the ProteinChip System, have been used to monitor different growth stages, to type strains, and to monitor drug resistance and effectiveness of drug treatment.
ProteinChip technology is currently in use at the National Cancer Institute, the National Institutes of Health (NIH), the Royal Free Hospital (UK), the John Innes Institute (UK), M. D. Anderson Hospital, the University of California-San Francisco Cancer Center, Stanford University, and Virginia Prostate Cancer Center.
Case Study 1: Prostate Cancer (Back to Top)
Validation of biomarkers by comparing the protein expression profiles in a large number of patients is an important step before clinical testing. At the recent American Association for Cancer Research Meeting in Philadelphia, Lance Liotta (NCI) and Emanuel Petricoin (FDA) announced their preliminary results using Ciphergen's ProteinChip System combined with Laser Capture Microdissection (LCM) to identify and partly validate several markers for prostate cancer (ref. 2). In this study, 500 to 1,000 cells were first obtained by LCM of frozen sections of tissue in normal, pre-cancerous, and cancerous stages. Following cell lysis, the sample was applied to the ProteinChip Array for binding, briefly washed, dried, and the protein mass profile was produced in the ProteinChip System. Several "phenomic fingerprints" were generated that revealed well-defined patterns of protein expression, which correlated to specific disease states. The ProteinChip Software allowed multiple data sets to be averaged, subtracted, combined, and compared to further elucidate specific changes in individual proteins.
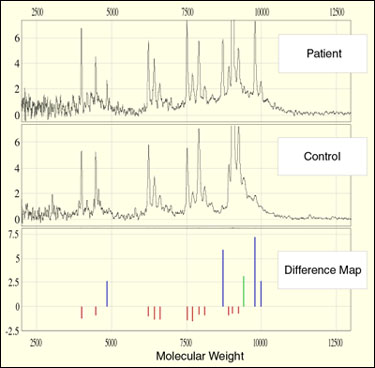
Case Study 2: Ovarian Cancer (Back to Top)
A similar approach was taken by Monica Brown (NCI) to identify biomarkers in ovarian cancer. At the recent 7th Biennial International Forum on Ovarian Cancer, Brown described her lab's approach to evaluate ovarian neoplasms by generating protein fingerprints using LCM-isolated ovarian cancer cells and the ProteinChip System (ref. 3). From 200 epithelial ovarian cancer cells from three moderately differentiated papillary serious tumors, they found a "phenomic fingerprint" with five to eight unique peptides of different molecular weights that were shared by all three ovarian tumors. Given the difficulty in early detection of ovarian cancer and rapid progression of the disease, this could be the first step in developing an effective diagnostic test and therapeutic interventions that could save many lives.
Case Study 3: Alzheimer's Disease (Back to Top)
Brian Austen's group at St. George's Hospital in London, studying Alzheimer's disease, has developed a ProteinChip assay to detect nanomolar quantities of beta amyloid peptides in biological fluids and tissue extracts. The beta amyloid biomarkers, thought to be at the root of the brain changes in Alzheimer's disease, were identified from only a few cells in a given sample. This research, which took only a week to complete, will enable studies of the cellular processing mechanisms of Amyloid Precursor Protein and could lead to a presymptomatic assay for Alzheimer's disease. An advantage of this ProteinChip assay over traditional technologies is the ability to develop an antibody-based assay for a peptide that simultaneously measures the presence of different beta amyloid peptide fragments quantitatively (ref. 4).
"In one week, we obtained robust data showing the up-regulation of several beta amyloid proteins in human cell lines exposed to high levels of cholesterol," said Austen. "We also obtained data showing down-regulation in cells treated with a cholesterol-lowering drug, lovastatin (Merck Research Laboratories). No other technique can analyze all the beta-amyloid variants with such confidence so rapidly. Recent research suggests that raised blood cholesterol levels induce deposits of the most neurotoxic forms of beta amyloid in the brain."
Seldi Protein Biology System II (Back to Top)
In May, Ciphergen launched the second emanation of its ProteinChip System, the Series PBS II, which consists of disposable ProteinChip array sets, ProteinChip reader, and Seldi Protocol Assistant software. ProteinChip array sets include chemically- and biochemically-enhanced ProteinChip arrays for specified applications, Seldi Protocol Assistant software and accessories defined for each application area.
- T.W. Hutchens and T.T Yip, "Surface Enhanced Neat Desorption (SEND): DNA bases capture of lactoferrin," Rapid Communication of Mass Spectrometry, 7: 576-580, 1993.
- C.P. Paweletz et al., "A novel, proteomic approach to monitor carcinogenic disease progression using surface enhanced desorption ionization spectroscopy (SELDI) of laser capture microdissection (LCM)-derived cells from cancer tissue," Proceedings of the American Association of Cancer Researchers, 40, March 1999.
- M.R. Brown, "The Application of Novel Technology to the Study of Ovarian Cancer," 7th Biennial International Forum on Ovarian Cancer—Stockholm, Sweden, 18-21 April 1999.
- B. Austen et al., "The role of cholesterol in the biosynthesis of beta-amyloid," Neuroreport 10 (8):1699-1705, 1999.
For more information: Lars Barfod, Ciphergen Biosystems Inc., 490 San Antonio Rd., Palo Alto, CA 94306. Tel: 650-496-3770.
By Laura DeFrancesco
